Author:
(1) Vojtech ˇ Cerm ˇ ak, Czech Technical University in Prague (Email: cermavo3@fel.cvut.cz);
(2) Lukas Picek, University of West Bohemia & INRIA (Email: picekl@kky.zcu.cz/lpicek@inria.cz);
(3) Luka´s Adam, Czech Technical University in Prague (Email: lukas.adam.cr@gmail.com);
(4) Kostas Papafitsoros, Queen Mary University of London (Email: k.papafitsoros@qmul.ac.uk).
Table of Links
- Abstract and Introduction
- Related work
- The WildlifeDatasets toolkit
- MegaDescriptor – Methodology
- Ablation studies
- Performance evaluation
- Conclusion and References
4. MegaDescriptor – Methodology
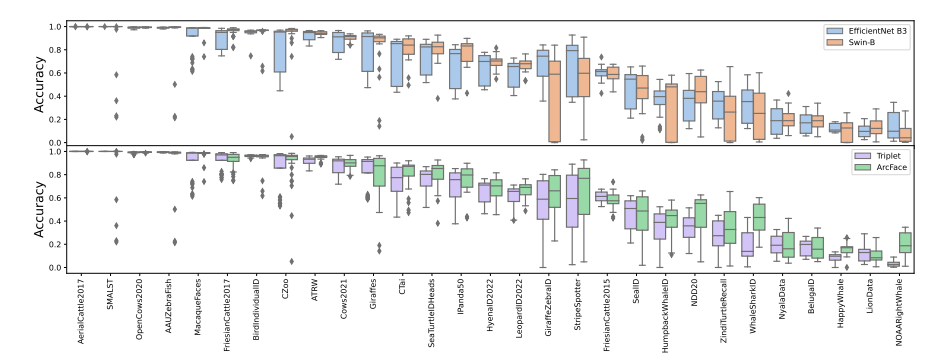
Wildlife re-identification is usually formulated as a closed-set classification problem, where the task is to assign identities from a predetermined set of known identities to given unseen images. Our setting draws inspiration from real-life applications, where animal ecologists compare a reference image set (i.e., a database of known identities) with a query image set (i.e., newly acquired images) to determine the identities of the individuals in new images. In the search for the best suitable methods for the MegaDescriptor, we follow up on existing literature [16, 31, 34, 41] and focus on local descriptors and metric Learning. We evaluate all the ablation studies over 29 datasets[4] provided through the WildlifeDataset toolkit.
4.1. Local features approaches
Drawing inspiration from the success of local descriptors in existing wildlife re-identification tools [21, 41], we include the SIFT and Superpoint descriptors in our evaluation. The matching process includes the following steps: (i) we extract keypoints and their corresponding descriptors from all images in reference and query sets, (ii) we compute the descriptors distance between all possible pairs of reference and query images, (iii) we employ a ratio test with a threshold to eliminate potentially false matches, with the optimal threshold values determined by matching performance on the reference set, and (iv) we determine the identity based on the absolute number of correspondences, predicting the identity with highest number from reference set.
4.2. Metric learning approaches
Following the recent progress in human and vehicle re-id [14, 35, 56], we select two metric learning methods for our ablation studies – Arcface [17] and Triplet loss [46] – which both learn a representation function that maps objects into a deep embedding space. The distance in the embedded space should depend on visual similarity between all identities, i.e., samples of the same individual are close, and different identities are far away. CNN- or transformer-based architectures are usually used as feature extractors.
The Triplet loss [26, 46] involves training the model using triplets (xa, xp, xn), where the anchor xa shares the same label as the positive xp, but a different label from the negative xn. The loss learns embedding where the distance between xa and xp is small while distance between xa and xn is large such that the former pair should be distant to latter by at least a margin m. Learning can be further improved by a suitable triplet selection strategy, which we consider as a hyperparameter. We consider ’all’ to include all valid triplets in batch, ’hard’ for triplets where xn is closer to the xa than the xp and ’semi’ to select triplets where xn is further from the xa than the xp.
The ArcFace loss [17] enhances the standard softmax loss by introducing an angular margin m to improve the discriminative capabilities of the learned embeddings. The embeddings are both normalized and scaled, which places them on a hypersphere with a radius of s. Value of scale s is selected as hyperparameter.
Matching strategy: In the context of our extensive experimental scope, we adopt a simplified approach to determine the identity of query (i.e., test) images, relying solely on the closest match within the reference set. To frame this in machine learning terminology, we essentially create a 1-nearest-neighbor classifier within a deep-embedding space using cosine similarity.
Training strategy: While training models, we use all 29 publicly available datasets provided through the WildlifeDataset toolkit. All datasets were split in an 80/20% ratio for reference and query sets, respectively, while preserving the closed set setting, i.e., all identities in the query set are available in the reference set. Models were optimized using the SGD optimizer with momentum (0.9) for 100 epochs using the cosine annealing learning rate schedule and mini-batch of 128.
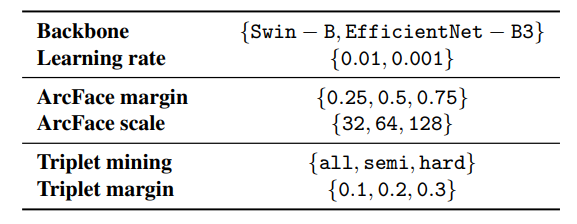
Hyperparameter tunning: The performance of the metric learning approaches is usually highly dependent on training data and optimization hyperparameters [35]. Therefore, we perform an exhaustive hyperparameters search to determine optimal hyperparameters with sustainable performance in all potential scenarios and datasets for both methods. Besides, we compare two backbone architectures – EfficientNet-B3 and Swin-B – with a comparable number of parameters. We select EfficientNet-B3 as a representative of traditional convolutional-based and Swin-B as a novel transformer-based architecture.
For each architecture type and metric learning approach, we run a grid search over selected hyperparameters and all the datasets. We consider 72 different settings for each dataset, yielding 2088 training runs. We use the same optimization strategy as described above. All relevant hyperparameters and their appropriate values are listed in Table 2.
This paper is available on arxiv under CC BY-NC-ND 4.0 DEED license.
[4] We avoided Drosophila (low complexity and high image number) and SeaTurleID2022 [5] due to its big overlap with SeaTurleIDHeads [38].

